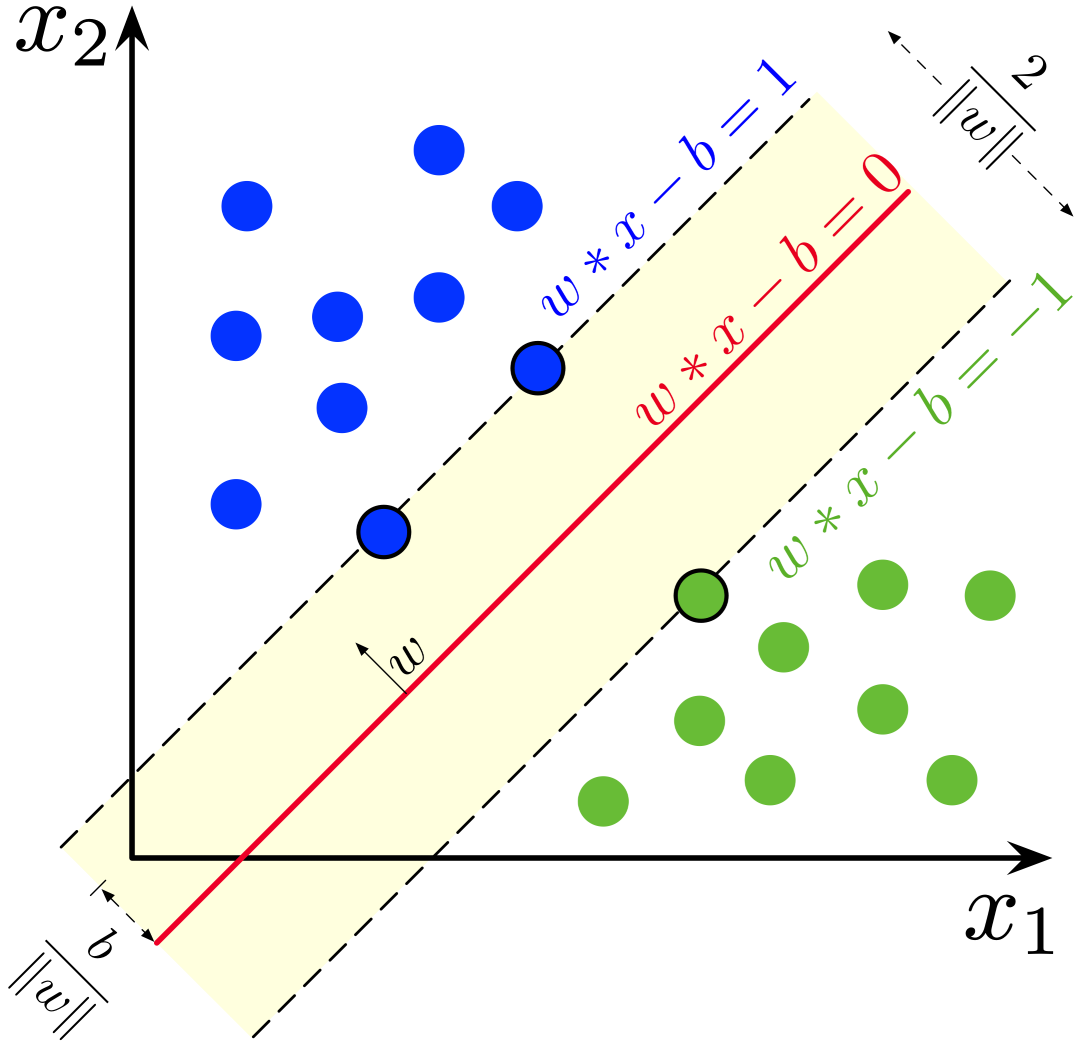
from sklearn.datasets import make_moons
from sklearn.model_selection import GridSearchCV
def nonlinear_svm_example():
# Generate non-linear dataset
X, y = make_moons(n_samples=1000, noise=0.15, random_state=42)
# Split the data
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
# Scale features
scaler = StandardScaler()
X_train_scaled = scaler.fit_transform(X_train)
X_test_scaled = scaler.transform(X_test)
# Create dictionary of SVMs with different kernels
kernels = ['linear', 'poly', 'rbf', 'sigmoid']
svm_classifiers = {
kernel: svm.SVC(kernel=kernel, random_state=42)
for kernel in kernels
}
# Train and evaluate each kernel
plt.figure(figsize=(20, 5))
for i, (kernel, classifier) in enumerate(svm_classifiers.items(), 1):
# Train classifier
classifier.fit(X_train_scaled, y_train)
# Make predictions
y_pred = classifier.predict(X_test_scaled)
# Create subplot
plt.subplot(1, 4, i)
# Plot training data
plt.scatter(X_train_scaled[y_train == 0, 0], X_train_scaled[y_train == 0, 1],
color='blue', marker='o', label='Class 0')
plt.scatter(X_train_scaled[y_train == 1, 0], X_train_scaled[y_train == 1, 1],
color='red', marker='s', label='Class 1')
# Create mesh grid
x_min, x_max = X_train_scaled[:, 0].min() - 0.5, X_train_scaled[:, 0].max() + 0.5
y_min, y_max = X_train_scaled[:, 1].min() - 0.5, X_train_scaled[:, 1].max() + 0.5
xx, yy = np.meshgrid(np.arange(x_min, x_max, 0.02),
np.arange(y_min, y_max, 0.02))
# Plot decision boundary
Z = classifier.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
plt.contourf(xx, yy, Z, alpha=0.4)
# Plot support vectors
plt.scatter(classifier.support_vectors_[:, 0], classifier.support_vectors_[:, 1],
s=100, linewidth=1, facecolors='none', edgecolors='k',
label='Support Vectors')
plt.title(f'{kernel.upper()} Kernel')
plt.xlabel('Feature 1')
plt.ylabel('Feature 2')
if i == 1:
plt.legend()
print(f"\n{kernel.upper()} Kernel Performance:")
print(classification_report(y_test, y_pred))
plt.tight_layout()
# plt.show()
def hyperparameter_tuning_example():
# Generate dataset
X, y = make_classification(n_samples=1000, n_features=2, random_state=42)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
# Scale features
scaler = StandardScaler()
X_train_scaled = scaler.fit_transform(X_train)
X_test_scaled = scaler.transform(X_test)
# Define parameter grid
param_grid = {
'C': [0.1, 1, 10],
'kernel': ['linear', 'rbf'],
'gamma': ['scale', 'auto', 0.1, 1],
'class_weight': [None, 'balanced']
}
# Create SVM classifier
svc = svm.SVC(random_state=42)
# Perform grid search
grid_search = GridSearchCV(
estimator=svc,
param_grid=param_grid,
cv=5,
n_jobs=-1,
scoring='accuracy'
)
grid_search.fit(X_train_scaled, y_train)
# Print results
print("Best parameters:", grid_search.best_params_)
print("Best cross-validation score:", grid_search.best_score_)
# Evaluate on test set
best_model = grid_search.best_estimator_
y_pred = best_model.predict(X_test_scaled)
print("\nTest Set Performance:")
print(classification_report(y_test, y_pred))
if __name__ == "__main__":
print("Running SVM Examples...")
print("\n1. Linear SVM Example:")
linear_svm_example()
print("\n2. Non-linear SVM Example:")
nonlinear_svm_example()
print("\n3. Hyperparameter Tuning Example:")
hyperparameter_tuning_example()